





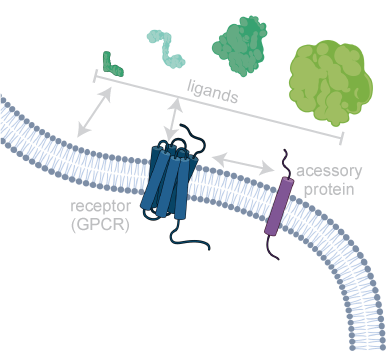
Membranes are the central interface between environment and cellular response. Consequently, membrane proteins are the most common targets for modern drugs. In addition, lipids themselves can directly modulate biological processes. We are investigating the molecular mechanisms underlying these central communication pathways, including membrane proteins (e.g. GPCRs), ligands (e.g. neuropeptides), and lipids. We are particularly interested in unraveling the role of the environment in shaping activity. To this end, we are developing and using innovative biochemical strategies and tailored biophysical experimental setups with the goal of gaining insight into previously inaccessible systems under increasingly native conditions.
Our research is driven by integrative approaches that combine the inherent strengths of solution and (DNP) solid-state NMR, fluorescence- and surface-based techniques, cryo-EM, microfluidics, and molecular dynamics simulations. This setup is essential to decipher the desired adequate picture of complex membrane systems.
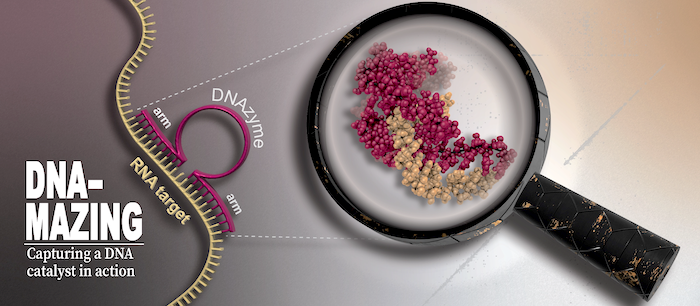
DNAzymes are high-precision biocatalysts capable of selectively eliminating unwanted RNA molecules. They consist of a short, single-stranded DNA sequence that can be divided into a fixed core region and two freely adaptable substrate binding arms. This modular design allows the development of specific variants for almost all conceivable target RNAs within a very short time by simply adapting the binding arms to the corresponding target RNA.
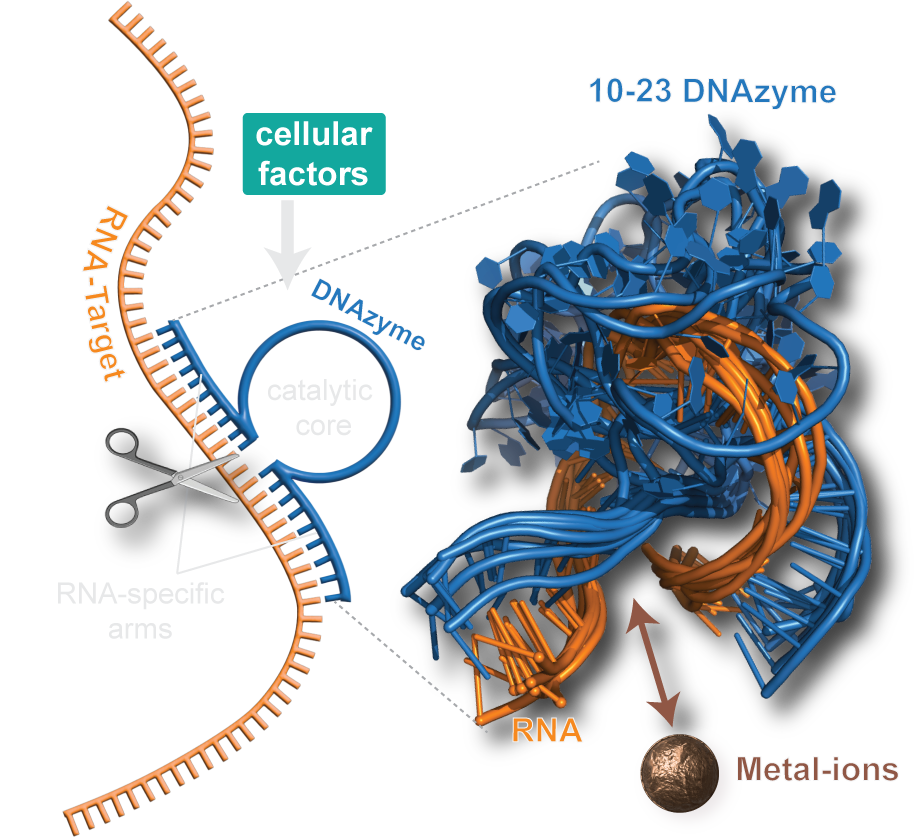
Unfortunately, the activity of classical DNAzyme variants is strongly reduced in the targeted cells, and randomized optimization strategies have not yet been able to resolve this dilemma. Recently, we could change this situation by providing long-needed mechanistic insights into one of the most popular DNAzyme variant. Our research highlights molecular features that are responsible for the low cellular activity and identifies possible chemical modifications that can resolve the persisting shortcomings of the DNAzyme technology. Our aim is to further increase the understanding of fundamental properties of different DNAzyme variants and simultanioulsy exploit our insights to develop a new generation of DNAzymes with increased bioactivity.
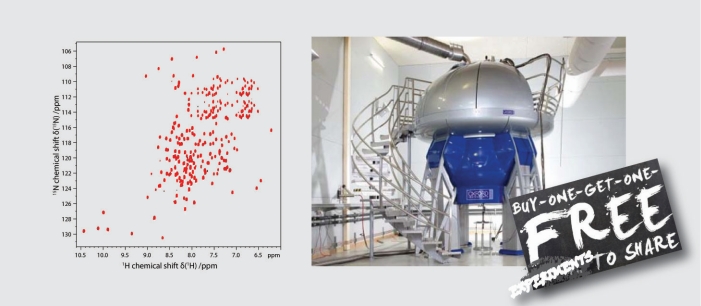
While NMR has the potential to obtain atomic-resolution insights into native biological systems, most NMR methods face increasing limitations when sample complexity is approaching native conditions. Our NMR method development therefore aims to achieve improvements that are particularly useful for challenging biological systems, including new pulse sequences and isotope labeling strategies. Our UTOPIA NMR setup allows e.g. the acquisition of relaxation favorable 13C and 15N spectra during the acquisition of the conventional 1H detected spectra. The additional spectra can be obtained for free and contain otherwise not accessible information.
UTOPIA NMR support: The current UTOPIA version is v0.9. Please contact us via email to receive current beta-versions of sequences and support. -Send Mail-
In general, NMR is also applicable directly in cellular environments. However, cellular NMR's limitations comprise mainly signal sensitivity as well as selectively of the protein of interest. Our targeted DNP approach combines the sensitivity boost of dynamic nuclear polarization (DNP) with the selectivity of protein interactions, facilitating the investigation of a target protein in a cellular context.